Research
 REGULATION OF GENE EXPRESSION DURING MUSCLE SPECIFICATION AND PATTERNING
REGULATION OF GENE EXPRESSION DURING MUSCLE SPECIFICATION AND PATTERNING
Myogenesis in Drosophila relies upon the activity of the Twist bHLH transcriptional
regulator during patterning of the embryonic mesoderm. Twist regulates a wide variety
of processes during mesodermal development, such as the specification and migration
of the mesoderm, segregation of
mesoderm into different domains, specification of the cells that will give rise to
the musculature, and morphogenesis of the final muscle pattern. Our goal is to understand
how a single transcription factor such as Twist can regulate such a diverse number
of processes during mesodermal development.
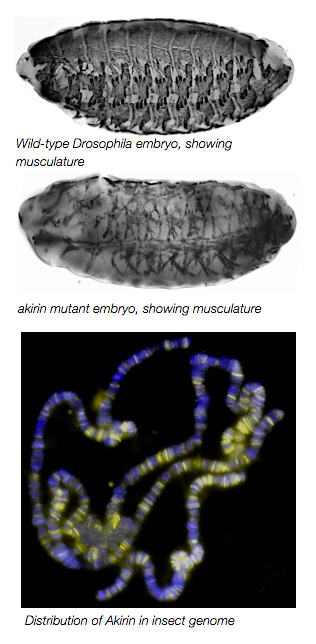
These different functions of Twist likely arise from interactions with other secondary activators. During my postdoctoral training in the lab of Dr. Mary Baylies at the Sloan-Kettering Institute, we identified the highly conserved nuclear protein Akirin as a novel cofactor of Twist at Twist-regulated enhancers. akirin mutants have severely disrupted muscle patterning and morphologies. Despite a lack of defined motifs or catalytic activity, we determined that Akirin is required for optimal expression of Twist-regulated genes that are critical for embryonic myogenesis. Mechanistically, we found that Akirin mediates a link between Twist transcription factor activity and the SWI/SNF-class Brahma chromatin remodeling complex to facilitate Twist-regulated transcription duringDrosophila embryonic development. These results suggest a new method for regulating SWI/SNF activity: the linking of transcription factor activities to chromatin remodeling factors by cofactors such as Akirin to facilitate tissue- and context-specific gene activation. The Nowak Lab is currently examining the molecular regulation of the Akirin protein and its interactions with Twist during embryonic development.
Nowak, S.J., Aihara, H., Gonzalez, K., Nibu, Y., and Baylies, M.K. 2012. PLoS Genet 8(3): e1002547
THE CELL BIOLOGY OF MYOBLAST FUSION
The final step of myogenesis, the fusion of myoblasts into a syncytial myotube, is the key event for formation and growth of muscle tissue. However, the cellular processes that occur, and the molecular machinery that regulates cell-cell fusion remain poorly understood. Studies of myoblast fusion in a variety of species, including mammals and Drosophila, indicated that the basic steps of myoblast fusion are highly conserved: Myoblasts migrate to and recognize their fusing partner, align their membranes, adhere to one another, and finally fuse to form a multinucleate structure. The kinetics of these events during mammalian myoblast fusion have not been well-defined. Further, while the molecular machinery that governed cell-cell fusion in Drosophila had been well-characterized, the gene products that regulated this process in mouse myoblasts remained unclear.
C2C12 myoblasts are a mouse myoblast cell line that can be easily manipulated to fuse into syncytial myotubes on demand. During my postdoctoral training in the Baylies Lab, we developed live imaging techniques to characterize the morphological events and cell-based behaviors that occur in C2C12 myoblasts as they proceed through differentiation and fusion. In tandem, we employed RNAi techniques to knock down the mouse orthologues of gene products that have been identified as critical for fusion ofDrosophila myoblasts. As an example, we found that regulators of Arp2/3-dependent actin cytoskeleton remodeling have a conserved role in both insect and mammalian myoblast fusion. Using our analysis of wild-type myoblast behaviors during the fusion process as a guide, we identified the specific step of the fusion process that requires these proteins. We conclude from these results that the cellular behaviors and molecular machinery that are critical for myoblast fusion are conserved across species from insects to mammals.
Nowak, S.J., Nahirney, P., Hadjantonakis, A-K., and Baylies, M.K. J Cell Sci. 2009 Sep 15;122(Pt 18):3282-93









