Research
My research centers around the mechanisms that control the activities of proteinsthat participate in cell signaling. I use evolutionary comparisons of protein sequences and molecular modeling to predict the parts of proteins most likely to control their activity. Then I test those predictions in the laboratory, using a combinationof techniques to perturb and measure activity. I investigate the structure, function and control of NADPH oxidases (NOX) and of voltage-gated proton channels (HV1). NOXes and HV1 are known to work together in several cell types, and both produce signals important not only to human health but also to the development, physiology, and lifestyles of many other organisms.
See below for what's been happening with current projects in the lab:
-
NADPH Oxidase(NOX) Structure, Function, and Control
Prokaryotic NOX
I use structural and sequence information to inform experiments, exploring different aspects of NADPH oxidase (NOX) structure and the isoform specific control of Nox activity, fundamental to understanding NOX's role as a signal generator. I am working with an international team -- notably Drs. Franck Fieschi and Marie Jose Stasia of Institut Biologie Structurale, along with many colleagues in Grenoble and Paris -- using a prokaryotic homolog, SpNOX, to learn some fundamental properties of NOXes in general; eventually what we learn should be useful towards drug discovery in human NOXes. Recently our large team characterized the full-length structure of SpNOX by SANS (Vermot et al, 2020, Biophys J. 119:605-618), the first full-length
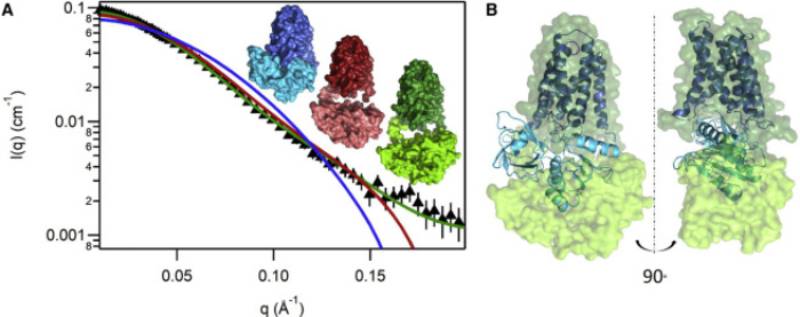 structural characterization of a NOX. From our results we posited that substrate
binding provides correct docking of the dehydrogenase domain onto the transmembrane
domain for electron transfer; this suggestion has been supported by later groundbreaking
cryo-EM structures from other groups. Our shared PhD student, now Dr. Annelise Vermot,
spearheaded a nice review of NOX structure, mechanism, and pathophysiology (Vermot et al, 2021, Antioxidants 10(6):890). Talented KSU undergrads Brittany Notice, Emily Onyekwere, James Scott, and Victoria
Gonzalez contributed mutagenesis, protein purification, and biochemical characterization
to this ongoing project, and lab manager Sarah Waldron was instrumental at every step.
structural characterization of a NOX. From our results we posited that substrate
binding provides correct docking of the dehydrogenase domain onto the transmembrane
domain for electron transfer; this suggestion has been supported by later groundbreaking
cryo-EM structures from other groups. Our shared PhD student, now Dr. Annelise Vermot,
spearheaded a nice review of NOX structure, mechanism, and pathophysiology (Vermot et al, 2021, Antioxidants 10(6):890). Talented KSU undergrads Brittany Notice, Emily Onyekwere, James Scott, and Victoria
Gonzalez contributed mutagenesis, protein purification, and biochemical characterization
to this ongoing project, and lab manager Sarah Waldron was instrumental at every step. Eukaryotic NOX
We used my molecular models to help understand phosphorylation of NOX2. (Beaumel et al. 2017. Free Rad. Biol. Med. 113:1-15). My models were used in the past in several other publications: (Chen, et al. 2015. Free Rad. Biol. Med. 89:793-805); (Dahan, et al. 2015. J. Leukoc. Biol. 98: 859-874) (Lorincz, et al. 2013. Free Rad. Biol. Med. 68, 65-71.)
Before coming to Kennesaw, I was involved in a project in the laboratory of Dave Lambeth at Emory University to discover small molecule inhibitors of NOXes, which could eventually be useful in treating diseases; others on the project included James McCoy, Yerun Zhu, Yang Li, Becky Diebold and Aiming Sun, and Shivaprakash Gangappa. The latest work from that project just appeared in print (De La Cruz et al, 2021, PLoSONE 16(7):e0254632). Older papers from this project are listed below:
- Li, Y., T. Ganesh, B.A. Diebold, Y. Zhu, J.W. McCoy, S.M.E. Smith, A. Sun, J. D. Lambeth. 2015. Thioxo-dihydroquinazolin-one Compounds as Novel Inhibitors of Myeloperoxidase. 6 (10):1047-1052.
- Diebold, B.A., S.M.E. Smith, Y. Li, J.D. Lambeth. 2014. NOX2 as a target for Drug Development: Indications, Possible Complications and Progress. Antioxidants and Redox Signaling. 23(5):375-405.
- Smith, S.M.E., J. Min, T. Ganesh, T. Kawahara, B. Diebold, Y. Zhu, A. Sun, J.P. Snyder, H. Fu, Y.Du, I. Lewis, J. D. Lambeth. 2012. Ebselen and congeners inhibit NADPH-oxidase 2 (Nox2)-dependent superoxide generation by interrupting the binding of regulatory subunits. Chem. Biol. 19, 752-763.
-
HV1 Proton Channel
Exploring pH sensing in HV1
 We mutated the Trp residue that appears in the signature sequence of HV1 (see Bioluminescence section below). Our electrophysiology colleagues in the DeCoursey
lab showed that these mutants affected several key properties. Trp mutants are the
first mutants of HV1 shown to change a hallmark property of HV1, its pH sensing. My molecular figure made the cover of JGP! See the image on the
right. (Cherny et al, 2015. JGP 146:343-356). Then Sarah Thomas, the Smith lab manager, cloned the gene for HV1 from snails. Again unexpectedly, the snail HV1 has impaired internal pH sensing, and we found a mutation in the human HV1 protein that recapitulated this property. (Thomas, et al 2018. J. Gen. Physiol 150:835-150) (Cherny, et al, 2018. J. Gen Physiol. 150:851-862)
We mutated the Trp residue that appears in the signature sequence of HV1 (see Bioluminescence section below). Our electrophysiology colleagues in the DeCoursey
lab showed that these mutants affected several key properties. Trp mutants are the
first mutants of HV1 shown to change a hallmark property of HV1, its pH sensing. My molecular figure made the cover of JGP! See the image on the
right. (Cherny et al, 2015. JGP 146:343-356). Then Sarah Thomas, the Smith lab manager, cloned the gene for HV1 from snails. Again unexpectedly, the snail HV1 has impaired internal pH sensing, and we found a mutation in the human HV1 protein that recapitulated this property. (Thomas, et al 2018. J. Gen. Physiol 150:835-150) (Cherny, et al, 2018. J. Gen Physiol. 150:851-862)HV1 Selectivity Filter -- it wanders, and the gasket helps it work
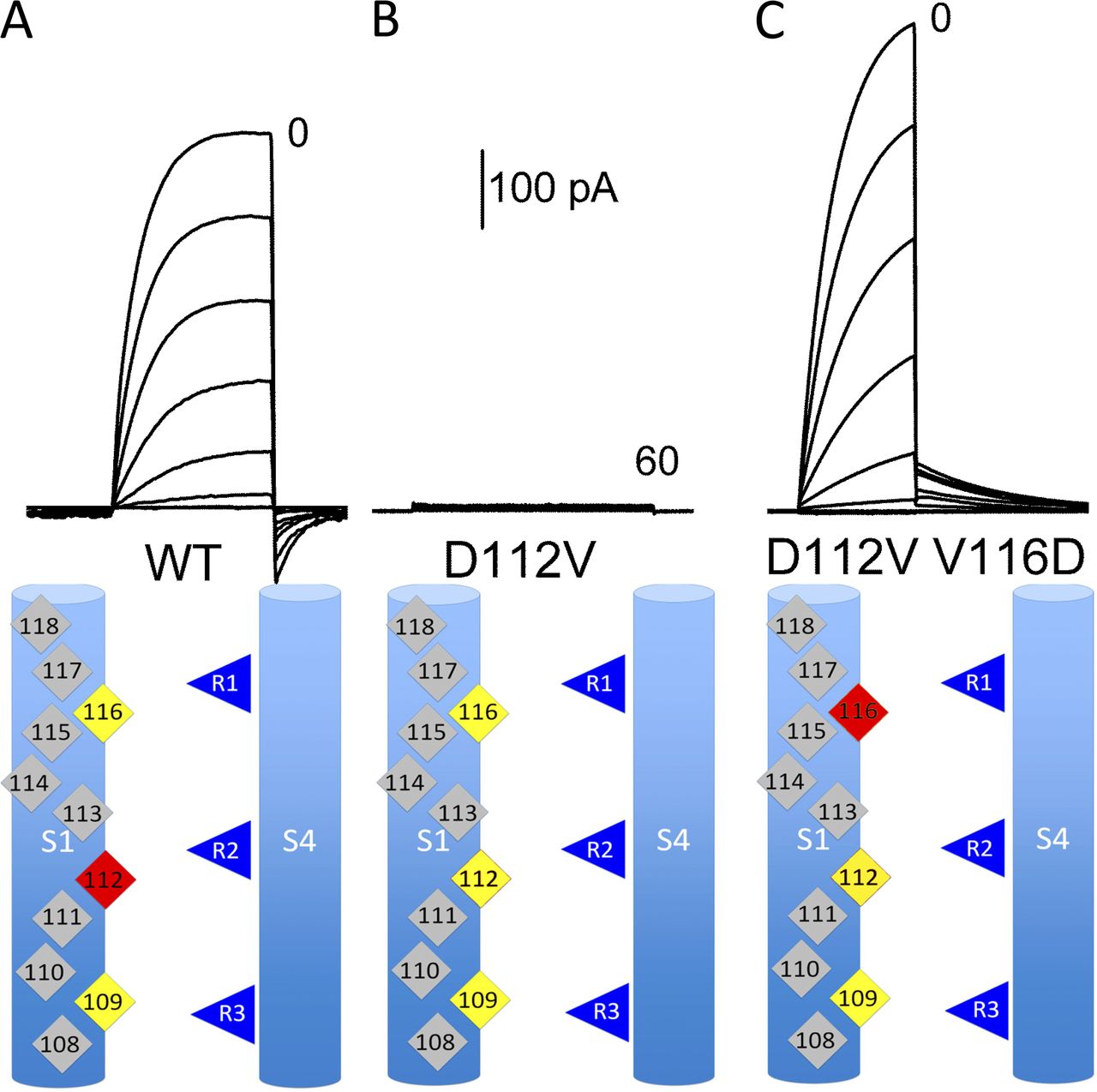 My sequence and phylogenetic analysis, combined with my molecular model, provided
a good template from which to make substitutions aimed at affecting proton conduction.
My sequence and phylogenetic analysis, combined with my molecular model, provided
a good template from which to make substitutions aimed at affecting proton conduction.The electrophysiology expertise of the DeCoursey lab enabled us to use these mutants to find (an essential part of) the selectivity filter of HV1, D112 in human HV1 (Musset et al, 2011 Nature 480:273-277). Surprisingly, most mutants at this site convert the perfectly proton selective HV1 to an anion selective channel. Also surprisingly, we can move the selectivity filter up one turn (but not down one turn) and retain the apparently perfect selectivity (Morgan et al 2013, JGP 142:625-40). Carmay Lim's group used a reduced model of the selectivity filter region that includes D112 and an arginine from S4 to perform molecular mechanics/quantum mechanics calculations that help explain how the proton moves through this area (Dudev et al 2015 Sci Reports 10320). Mutations in the 'hydrophobic gasket' that surrounds this region affect selectivity and other important properties of the channel (Banh et al, 2019, PNAS 116:18951-18961), supporting the idea that D112 does not act alone to provide selectivity.
HV1 Molecular Model helps us understand HV1 function
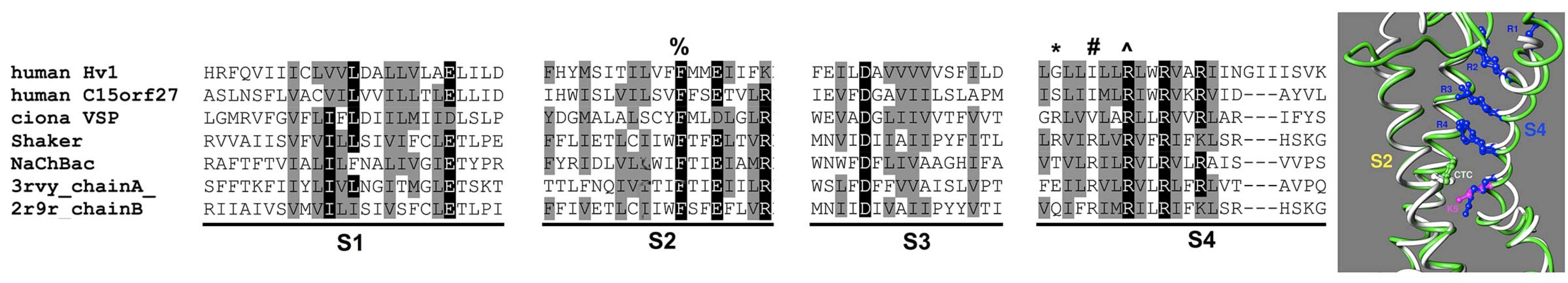
Modeling of voltage sensor domains (VSD) is tricky because the mobility of the S4 helix allows different alignments of the 'arginine registry' depending on the functional state of the sensor. I used phylogenetic analysis and a structurally informed alignment method to fix the registry to the open state, and then Kethika Kullepumera in Regis Pomes's lab tested two alternative homology models with massively repeated molecular dynamics simulations. The preferred model from this analysis produced a hypothesis about the accessibility of the third arginine on S4, which when tested via patch clamp in Tom DeCoursey's lab turns out to be accessible to the solution on the inside of the membrane, even in the open state (Kulleperuma et al 2013, JGP 141:445-465). Most recently, we used the serendipitous high affinity Zn binding effect of a mutant to show that voltage sensor rearrangements during opening move the first arginine on S4 relative to residues on S1 and S2 (Cherny et al, 2020, JGP 152 (10), e202012664).
HV1 and Bioluminescence
I developed signature sequences to predict HV1’s in other organisms. With Al Place’s help, we found a putative HV1 gene in a dinoflagellate cDNA library, now confirmed as a bona fide HV1 protein by patch clamping in Tom DeCoursey's lab. (Smith et al 2011, PNAS 108: 18162–18167). Then Juan Rodriguez, a MSIB student in my lab, helped prove that HV1 in Lingulodinium polyedrum is right where Woody Hastings predicted it would be! (Rodriguez et al, 2017, PLoSOne journal.pone.0171594). My undergraduate Gabe Kigundu showed that HV1 is expressed in many non-bioluminescent dinoflagellates, including Oxyrrhis marina which is taken to be basal to dinoflagellates, indicating that HV1 probably has some other conserved function in dinoflagellates (Kigundu et al 2018, J. Euk. Microbiol. 65 (6): 928-933).
Here's a link about our discovery of a dinoflagellate proton channel gene, with an illustration of how we think it works to trigger bioluminescence:
NSF.gov story -
Biomarkers for post-operative deliriumFrom discussions following Dr. Doreen Wagner's excellent seminar at the John Salerno Memorial Symposium came a new interdisciplinary grant funded by the American Association of Critical Care Nurses (Dr. Wagner, PI), to explore connections between perioperative hypothermia, postoperative delirium, and biomarkers of inflammation and oxidative stress. A team of nurses at Wellstar Kennestone hospital will consent the patients, assess them for delirium, and take blood samples. The lab team -- Dr. Wagner, Dr. Sharon Pearcey of Psychology, four KSU Biology undergrads, and I -- will perform ELISAs to quantitate the biomarkers in the patient blood. Then Dr. Pearcey will lead us on the data analysis to determine any relationships between clinical data and biomarker data. It's my first clinical project -- challenging but fun!
Learn more about RESEARCH in the College of Science and Mathematics at Kennesaw State University.








