Research
Enzymes are nature’s powerful, selective biochemical catalysts. Your body relies continuously and utterly on its enzymes to function properly. Likewise, many illnesses are directly related to the function of specific enzymes. Modern medicine takes great advantage of this situation, using designed inhibitors of specific enzymes to treat a wide variety of diseases and disorders.
For the chemist, the greatest value of enzymes comes from their highly evolved structures. Enzymes can tell us what optimal reaction structures look like and, if we are persistent, how those structures operate. Such knowledge could be exploited in the development of new catalysts or for mechanistic understanding in general, or in the development of more effective medicines. My lab is working to improve the fundamental understanding of organic structure and mechanism. Our main focus is on long-range effects in protein strands and enzyme complexes.
In 2004, we published a paper on extensive alignments of atomic orbitals that we recognized in natural complexes of trypsin and subtilisin. The alignments imply to us that each of these serine proteases can exploit a kind of long-range polarizability in its chemical dynamics. In the same paper, we described a simple non-enzymatic solvolysis reaction of oligopeptides. Its kinetics data plainly showed strong long-range effects of structure in reacting amino acid sequences.
Several students have helped to identify basic factors behind the observed long-range effects. Their work has involved the synthesis of many novel amino acid and oligopeptide derivatives for reactivity analysis. As a result, it is now clear that crowding, conformation, and stereochemistry along a strand can all contribute significantly to reactivity at other specific sites in the strand. As we identify additional basic factors, we can also begin to ask how Nature uses these factors to control and optimize an enzyme's catalytic power.
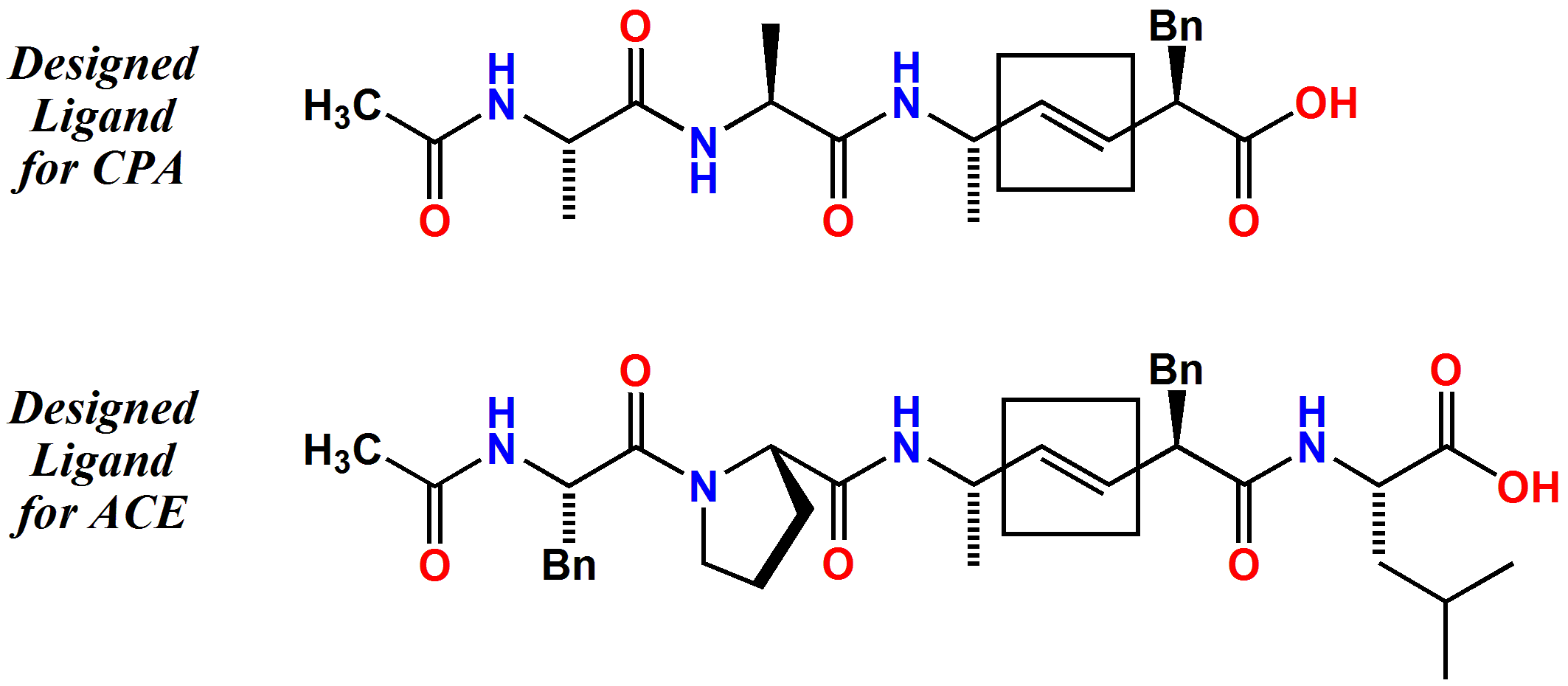
Some students have been preparing substrate-like ligands for other enzymes. Our systems of interest include the metalloproteases carboxypeptidase A and angiotensin-converting enzyme. Carboxypeptidase A, or 'CPA', is involved in digestion, post-translational modification of proteins, blood clotting, and reproduction. Angiotensin-converting enzyme, or 'ACE', helps to regulate blood pressure by the constriction of blood vessels. Inhibitors of ACE are used medicinally to control blood pressure. Our initial goal for each of these enzymes is to better characterize their complexes in terms of geometric detail at the active site. Our designed ligands contain alkene isosteres of the peptide bond. This means that the molecules are chemically resistant to enzyme action while resembling very closely the shape of a protein strand. Complexes between the ligands and the enzymes can then show us how each enzyme normally grips its natural targets, and therefore how atoms and electrons can move during the enzymatic chemistry.
Students on these projects have carried out conformational analysis of enzyme systems, multi-step synthesis of oligopeptides and related compounds, and reaction kinetics monitored by NMR.








