Research Interests
My lab is interested in understanding how the nervous system forms and functions, and what happens when things go wrong in the process. We primarily use the nematode Caenorhabditis elegans as a mode for these studies. The “worm” is a powerful system for looking at the nervous system, with the powerful transgenic tools and genetic approaches available to investigate gene function during development. We are particularly interested in understanding how worm orthologs of human disease genes function and what goes wrong when those genes are mutated, with the hope that lessons learned in the worm can be applied back to understanding human neurological disorders such as Autism Spectrum Disorder and schizophrenia.
The lab has two main research directions:
Sensory circuit assembly and function
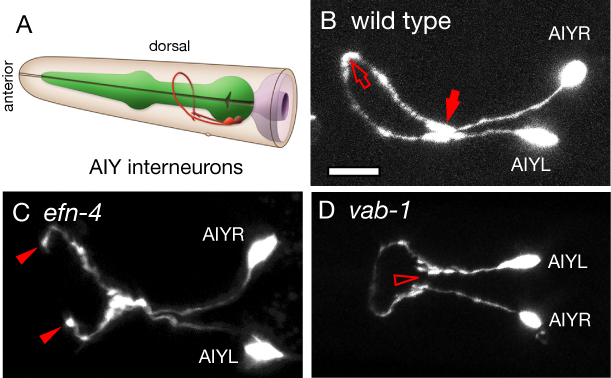
What genes are required to connect one neuron with another, what happens when this goes wrong, and what are the behavioral consequences of those defects? Previous work from the lab showed that the ephrin gene efn-4 is required for axon outgrowth in the AIY interneurons. These are the first layer of interneurons required for sensory signal processing from thermosensory and chemosensory neurons. Although efn-4 mutants look broadly normal, we have identified defects in their ability to identify and process chemosensory information. We are currently using synaptic markers to investigate how the various neurons in this circuit connect, and the role for efn-4 in this process.
This project, “Interneuron shape and sensory circuit robustness” was funded by NIH R15 grant NS100632-01 (September 2016 – August 2020, $378,561, PI: Hudson)
Schwieterman, A. A.*, Steves, A. N.+*, Yee, V. *, Donelson, C. J.+, Bentley, M. R. +, Santorella, E. M. +, Mehlenbacher, T.+, Pital, A.+, Howard, A. M.+, Wilson, M. R.+, Ereddia, D. E.+, Effrein, K. S.+, McMurry, J. L., Ackley, B. D., Chisholm, A. D., and Hudson, M. L. (2016). The C. elegans ephrin EFN-4 functions non-cell autonomously with heparan sulfate proteoglycans to promote axon outgrowth and branching. Genetics, 202, 639-660. (*These authors contributed equally to this work. +Undergraduate researcher). PMID: 26645816
Using comparative transcriptomics to understand proneural gene function
Gene regulatory networks dictate the proper establishment and function of the nervous system. Defects in gene regulatory systems have been implicated in multiple polygenic neurological conditions, including schizophrenia and autism spectrum disorders (ASDs). Of critical importance are the temporally ordered expression and subsequent regulation of transcription factors directing neurogenesis, neural migration, terminal fate, and target selection. While much is known about the transcription factors required to “switch on” the terminal fate of a neuron (“terminal selector” transcription factors), less is known about how upstream “proneural” transcription factors work during this process.
We have developed whole embryo comparative transcriptomics as an approach to identifying downstream targets of proneural genes. The single cell RNAseq dataset from John Murray’s lab can be used to identify candidate proneural transcription factors (Packer et al. 2019). We then isolate embryonic mRNA from worm strains bearing mutations in candidate proneural transcription factor genes then compare that against wild type embryonic mRNA to identify which transcripts go up or down in expression. Proneural transcription factor targets can then be validated using genetically encoded fluorescent reporter genes. Using this approach, we identified ceh-5 as a downstream target of cnd-1/NeuroD1 (Aquino-Nunez et al. 2020). We also identified eight downstream transcription factors as candidate targets of ngn-1/neurogenin (Christensen et al. 2020).
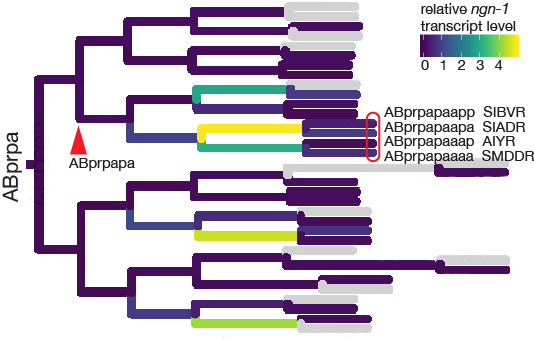
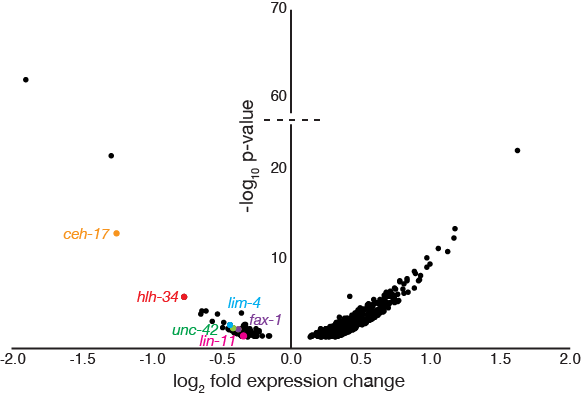
This project, “Identifying the gene regulatory network of neurogenin” is funded by NIH R15 grant GM140472-01 (October 2020 – September 2023, $406,500, PI: Hudson)
Aquino-Nunez, W.*, Mielko, Z. E.*, Dunn, T.+, Santorella, E. M.+, Hosea, C.+, Leitner, L.+, McCalla, D.+, Simms, C.+, Verola, W. M.+, Vijaykumar, S.+ and Hudson, M. L. (2020). cnd-1/NeuroD1 Functions with the Homeobox Gene ceh-5/Vax2 and Hox Gene ceh-13/labial To Specify Aspects of RME and DD Neuron Fate in Caenorhabditis elegans. Genes|Genomes|Genetics, 10, 3071-3085. PMID: 32601060. (*These authors contributed equally to this work. +Undergraduate researcher).
Christensen, E. L., Beasley, A.+, Radchuk, J.+, Mielko, Z. E., Preston, E., Stuckett, S., Murray, J. I. and Hudson, M. L. (2020) ngn-1/neurogenin activates transcription of multiple terminal selector transcription factors in the Caenorhabditis elegans nervous system. 2020. Genes|Genomes|Genetics, 10, 1949-1962. (+Undergraduate researchers). PMID: 32273286








